结直肠癌(Colorectal cancer,CRC)是全球流行的恶性肿瘤之一,全球癌症统计2020结果显示结直肠癌的发病率位于第三位,死亡率仅次于肺癌位居第二[1]。中国人的健康也承受着结直肠癌的威胁,其发病率和死亡率逐年增长[2]。随着治疗方案的进步,结直肠癌的生存率略有提高,但是受益人群有限[3, 4]。因此,研究结直肠癌的肿瘤微环境(tumor microenvironment, TME),尤其是免疫微环境,具有重要的意义。不同分区结直肠癌具有不同的特征,对传统放化疗、靶向治疗以及免疫治疗的疗效也存在差异[5-10]。因此,探索不同分区之间结直肠癌的差异有一定的临床意义。
结直肠癌具有复杂的肿瘤微环境,对结直肠癌的进展和预后有重要的影响。传统的高通量测序为结直肠癌的肿瘤微环境研究带来了进展,但无法准确反映肿瘤细胞的异质性。单细胞测序技术帮助我们进一步在单个细胞水平上对结直肠癌进行研究[11-13]。基于此技术,肿瘤免疫微环境得到更广泛的研究,一定程度上剖析了T细胞和B细胞的特征以及相互作用[14-16]。近年来在多种肿瘤中发现B细胞以及三级淋巴结构与肿瘤的良好预后相关并且可以预测免疫治疗的疗效[14, 17-19]。然而,B细胞在对预测结直肠癌的预后这一观点尚有争议,其组成和作用有待进一步研究。
该论文通过整合自测数据和GSE132465数据集,对来自CRC四个区域的19例样本的单细胞数据进行了联合分析,经过质控和去批次效应后,共获得39484个细胞进行下游分析,注释得到6种细胞类型。进一步对各区域内肿瘤组织中免疫细胞内部各亚型的浸润情况分析,发现从近端到远端CRC中, B细胞呈减少趋势。肿瘤组织中的浆细胞由正常组织中的IgA表型转变为IgG表型。不同区域中的B细胞在分化状态上存在差异。联合TCGA数据分析发现,CD20+ B细胞是B细胞中的重要群体,能够预测CRC患者的预后,并且与CD3+ T细胞相互作用,共同影响CRC患者的总生存期。最后,通过同源小鼠皮下移植瘤模型和患者来源的类器官模型验证CD20+ B细胞在肿瘤生长中起着重要作用,并抑制了抗PD-1的肿瘤杀伤作用。
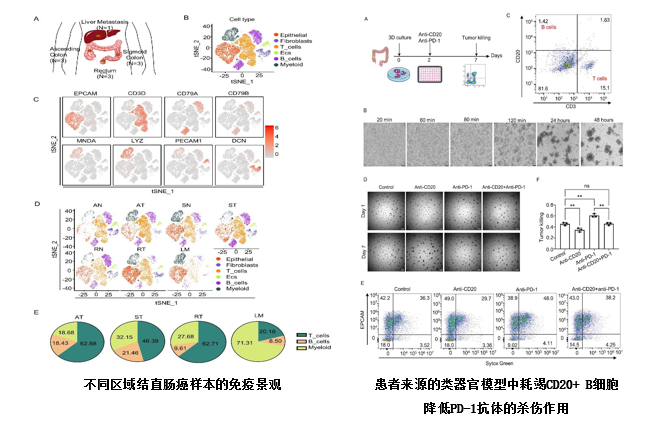
该研究展示了来自不同区域的CRC的免疫特征,并且发现CD20+ B细胞浸润与CRC患者的预后有关并可能促进抗PD-1的肿瘤杀伤作用,有望给结直肠癌的免疫治疗提供新的靶点。
南京大学附属金陵医院肿瘤科褚晓源教授、雷增杰副教授、傅恭博副教授,西南医院肿瘤科周杰副教授为该论文共同通讯作者,南京大学附属金陵医院纪琳琳硕士、黄梦茜博士、考晓明博士、朱家龙硕士为论文的共同第一作者。
原文链接:
https://doi.org/10.1016/j.canlet.2024.216664
参考文献:
[1] H. Sung, J. Ferlay, R.L. Siegel, et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA Cancer J Clin, 71 (2021) 209-249.
[2] W. Chen, R. Zheng, P.D. Baade, et al., Cancer statistics in China, 2015, CA Cancer J Clin, 66 (2016) 115-132.
[3] E. Dekker, P.J. Tanis, J.L.A. Vleugels, et al., Colorectal cancer, The Lancet, 394 (2019) 1467-1480.
[4] J.F. Shi, L. Wang, J.C. Ran, et al., Clinical characteristics, medical service utilization, and expenditure for colorectal cancer in China, 2005 to 2014: Overall design and results from a multicenter retrospective epidemiologic survey, Cancer-Am Cancer Soc, 127 (2021) 1880-1893.
[5] M.S. Lee, D.G. Menter, S. Kopetz, Right Versus Left Colon Cancer Biology: Integrating the Consensus Molecular Subtypes, J Natl Compr Canc Netw, 15 (2017) 411-419.
[6] R. Dienstmann, Tumor Side as Model of Integrative Molecular Classification of Colorectal Cancer, Clin Cancer Res, 24 (2018) 989-990.
[7] J.M. Loree, A.A.L. Pereira, M. Lam, et al., Classifying Colorectal Cancer by Tumor Location Rather than Sidedness Highlights a Continuum in Mutation Profiles and Consensus Molecular Subtypes, Clin Cancer Res, 24 (2018) 1062-1072.
[8] J. Guinney, R. Dienstmann, X. Wang, et al., The consensus molecular subtypes of colorectal cancer, Nat Med, 21 (2015) 1350-1356.
[9] B. Baran, N.M. Ozupek, N.Y. Tetik, et al., Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature, Gastroenterol Res, 11 (2018) 264-273.
[10] S.Y. Yang, M.S. Cho, N.K. Kim, Difference between right-sided and left-sided colorectal cancers: from embryology to molecular subtype, Expert Rev Anticancer Ther, 18 (2018) 351-358.
[11] J. Eberwine, J.Y. Sul, T. Bartfai, et al., The promise of single-cell sequencing, Nat Methods, 11 (2014) 25-27.
[12] E. Pennisi, Chronicling embryos, cell by cell, gene by gene, Science, 360 (2018) 367.
[13] E. Hedlund, Q. Deng, Single-cell RNA sequencing: Technical advancements and biological applications, Mol Aspects Med, 59 (2018) 36-46.
[14] W. Wang, Y. Zhong, Z. Zhuang, et al., Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer, Clin Transl Med, 11 (2021) e253.
[15] Y. Mei, W. Xiao, H. Hu, et al., Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer, Clin Transl Med, 11 (2021) e422.
[16] Y. Liu, Q. Zhang, B. Xing, et al., Immune phenotypic linkage between colorectal cancer and liver metastasis, Cancer Cell, 40 (2022) 424-437 e425.
[17] R. Cabrita, M. Lauss, A. Sanna, et al., Tertiary lymphoid structures improve immunotherapy and survival in melanoma, Nature, 577 (2020) 561-565.
[18] B.A. Helmink, S.M. Reddy, J. Gao, et al., B cells and tertiary lymphoid structures promote immunotherapy response, Nature, 577 (2020) 549-555.
[19] F. Petitprez, A. de Reynies, E.Z. Keung, et al., B cells are associated with survival and immunotherapy response in sarcoma, Nature, 577 (2020) 556-560.